Benchmark
Here we compare the performance of Naive Discrimination Learning in pyndl with other implementations, namely from the R packages ndl and ndl2.
This document summarizes the benchmarking procedure and results; the code to run these is available in the repository’s benchmark folder.
Creating the event files
The benchmark datasets are trigram-to-word events on the text of the
German Graphem Wikipedia page, that can be generated with python preprocessing.py.
Cues: ['aph', 'em#', 'hem', 'gra', 'rap', '#gr', 'phe'], Outcome: ['graphem']
Cues: ['aus', '#au', 'us#'], Outcome: ['aus']
Cues: ['wik', '#wi', 'kip', 'iki', 'edi', 'ia#', 'ped', 'ipe', 'dia'], Outcome: ['wikipedia']
…
Cues: ['ქარ', '#ქა', 'ართ', 'ლი#', 'რთუ', 'თულ', 'ული'], Outcome: ['ქართული']
Cues: ['ша#', 'қша', 'ақш', '#қа', 'зақ', 'қаз', 'аза'], Outcome: ['қазақша']
Cues: ['#한국', '한국어', '국어#'], Outcome: ['한국어']
…
The text contains some non-ASCII characters from various languages. While this is not a problem with pyndl, these characters would crash ndl2. This is why create cue-outcome events with a basic preprocessing, including just ASCII characters and letters. Please note, that in research applications we recommend using dedicated software for more careful preprocessing.
preprocess.create_event_file(corpus_file=txt_file,
event_file=ascii_event_file,
allowed_symbols='a-zA-Z0-9',
context_structure='document',
event_structure='consecutive_words',
event_options=(1,), # number of words,
cue_structure="trigrams_to_word",
lower_case=True,
remove_duplicates=True)
Based on this event file, we construct large scale event file by concatenating all events multiple times.
ascii_events = list(io.events_from_file(ascii_event_file))
io.events_to_file(ascii_events * times, event_dir / f"{times}_times_{ascii_event_file.name}", compatible=True)
Running pyndl
Our benchmark compares the runtime of pyndl’s NDL implementations on event files of varying size.
The naive dict_ndl learner is orders of magnitude slower and thus excluded from the comparison.
In addition to single processing, both parallel processing methods
(threading and OpenMP) are included. Run the benchmark with python run_ndl.py, which
saves the wall-clock times in a CSV files.
def clock(func, args, **kwargs):
gc.collect()
start = time.time()
result = func(*args, **kwargs)
stop = time.time()
duration = stop - start
return result, duration
…
for r in range(repeats):
for file_path in event_dir.glob('*.tab.gz'):
_, duration_omp1 = clock(ndl.ndl, (file_path, ALPHA, BETAS, LAMBDA_), n_jobs=1, method='openmp')
_, duration_omp4 = clock(ndl.ndl, (file_path, ALPHA, BETAS, LAMBDA_), n_jobs=4, method='openmp')
…
Running R ndl and ndl2
The R benchmark includes the equilibrium learner of ndl and
the iterative learner of ndl2. R -f run_ndl.R runs the ndl2 on the same event files as pyndl,
while ndl only runs on the smaller files because it is orders of magnitude slower.
ndl can be installed from CRAN by running install.packages('ndl') in R,
while ndl2 needs to be downloaded from Github first (ndl2, install with R CMD INSTALL ndl2.tar.gz).
Both ndl and ndl2 cannot handle compressed event files right away. For ndl, we load the full event file into memory (included in time measurement), while ndl2 reads an uncompressed event files.
…
for (r in 1:10) {
file_path <- event_files[i]
for (i in 1:n) {
st_proc_time <- system.time({
learner <- learnWeightsTabular(gsub("[.]gz$", "", file_path), alpha=0.1, beta=0.1, lambda=1.0, numThreads=1, useExistingFiles=FALSE)
})
mt_proc_time <- system.time({
learner <- learnWeightsTabular(gsub("[.]gz$", "", file_path), alpha=0.1, beta=0.1, lambda=1.0, numThreads=4, useExistingFiles=FALSE)
})
…
if (nrow(event_df) < 100000) {
proc_time <- system.time({
event_df <- read.delim(gzfile(file_path))
output <- estimateWeights(cuesOutcomes=event_df)
}, gcFirst = TRUE)
}
…
}
…
}
Results and discussion
We visualize the wall-clock time for the various NDL implementations and number of events as a line
plot with error bars for the standard-error within the ten repetitions (python plot_result.py, requires the plotting packages seaborn).
The shown wall-clock times were recorded on a laptop (Intel(R) Core(TM) i7-8565U CPU running Ubuntu 20.4, R 3.6 and python 3.9).
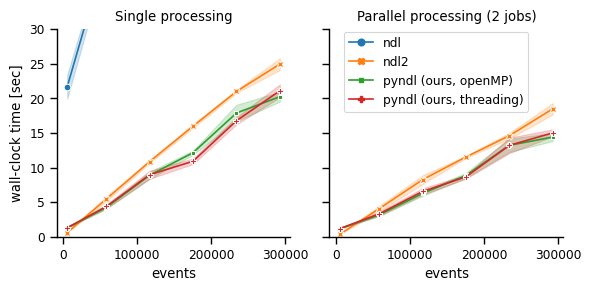
For small event files the pyndl are less than one second slower than ndl2 but still almost twenty times faster than ndl. With increasing number of events, pyndl becomes the fastest method in both single and parallel processing (2 jobs).
pyndl processes the event file into a faster accessible format, which results in the overhead for event files. This is similarly done in ndl2 where this overhead for small event files seems to be less time consuming. However, in contrast to the implementations in ndl2 and ndl, the implementation in pyndl never reads the full event file into memory, which is faster and has a smaller memory footprint than its competitors.